Podospora anserina Genome Project
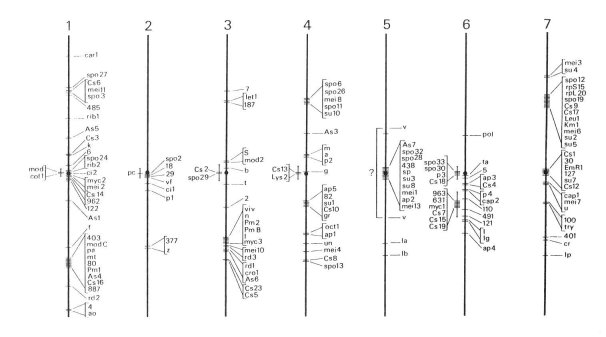
Podospora anserina is used over 60 years for genetics studies and a detailed genetic map is vailable (Marcou et al., 1993 in Genetic Maps, 6th Ed.).
These early genetic studies showed that P. anserina genome is distributed on 7 nuclear chromosomes (labelled LG I to LG VII) and one mitochondrial chromosome. Gel electrophoresis has allowed to accurately measure the genome size. P. anserina, like all filamentous fungi, has a small genome (circa 35 millions base pairs). With current assembly genome size is estimated at 35-36 megabases.
During the 1980's, the complete sequence of the mitochondrial genome of the s ("small s") strain has been established. It can be downloaded from the NCBI.
The complete sequence project started with a pilot project aimed to establish the sequence of the region bordering the centromere of LG V. Sequences can be downloaded at NCBI or EMBL with Accession #: BX088700 and AL627362.
Analysis of the region sequenced during the pilot project shows that the genes contain 1-2 introns, which have a small size (about 60 nucleotides). There is 1 kb in average between each gene. Results of the analysis are published in Fungal Genetics and Biology : Philippe Silar, Christian Barreau, Robert Debuchy, Sébastien Kicka, Béatrice Turcq, Annie Sainsard-Chanet, Carole H. Sellem, Alain Billault, Laurence Cattolico, Simone Duprat and Jean Weissenbach (2003) Characterization of the genomic organization of the region bordering the centromere of chromosome V of Podospora anserina by direct sequencing, Volume 39 n°3 Pages 250-263
A first draft of the genome was established in collaboration with Génoscope and the paper presenting the major features of the P. anserina genome is freely available in Genome Biology (direct link to the paper)
A finishing of the genome of strain S mat+ is now underway. Génoscope has produced a new assembly with a 20X coverage of 454 sequences. The successive Releases presented on this site stem from a combination of the old and new assemblies as well as manual targeted gap filling by PCR.
 |
These early genetic studies showed that P. anserina genome is distributed on 7 nuclear chromosomes (labelled LG I to LG VII) and one mitochondrial chromosome. Gel electrophoresis has allowed to accurately measure the genome size. P. anserina, like all filamentous fungi, has a small genome (circa 35 millions base pairs). With current assembly genome size is estimated at 35-36 megabases.
During the 1980's, the complete sequence of the mitochondrial genome of the s ("small s") strain has been established. It can be downloaded from the NCBI.
The complete sequence project started with a pilot project aimed to establish the sequence of the region bordering the centromere of LG V. Sequences can be downloaded at NCBI or EMBL with Accession #: BX088700 and AL627362.
Analysis of the region sequenced during the pilot project shows that the genes contain 1-2 introns, which have a small size (about 60 nucleotides). There is 1 kb in average between each gene. Results of the analysis are published in Fungal Genetics and Biology : Philippe Silar, Christian Barreau, Robert Debuchy, Sébastien Kicka, Béatrice Turcq, Annie Sainsard-Chanet, Carole H. Sellem, Alain Billault, Laurence Cattolico, Simone Duprat and Jean Weissenbach (2003) Characterization of the genomic organization of the region bordering the centromere of chromosome V of Podospora anserina by direct sequencing, Volume 39 n°3 Pages 250-263
A first draft of the genome was established in collaboration with Génoscope and the paper presenting the major features of the P. anserina genome is freely available in Genome Biology (direct link to the paper)
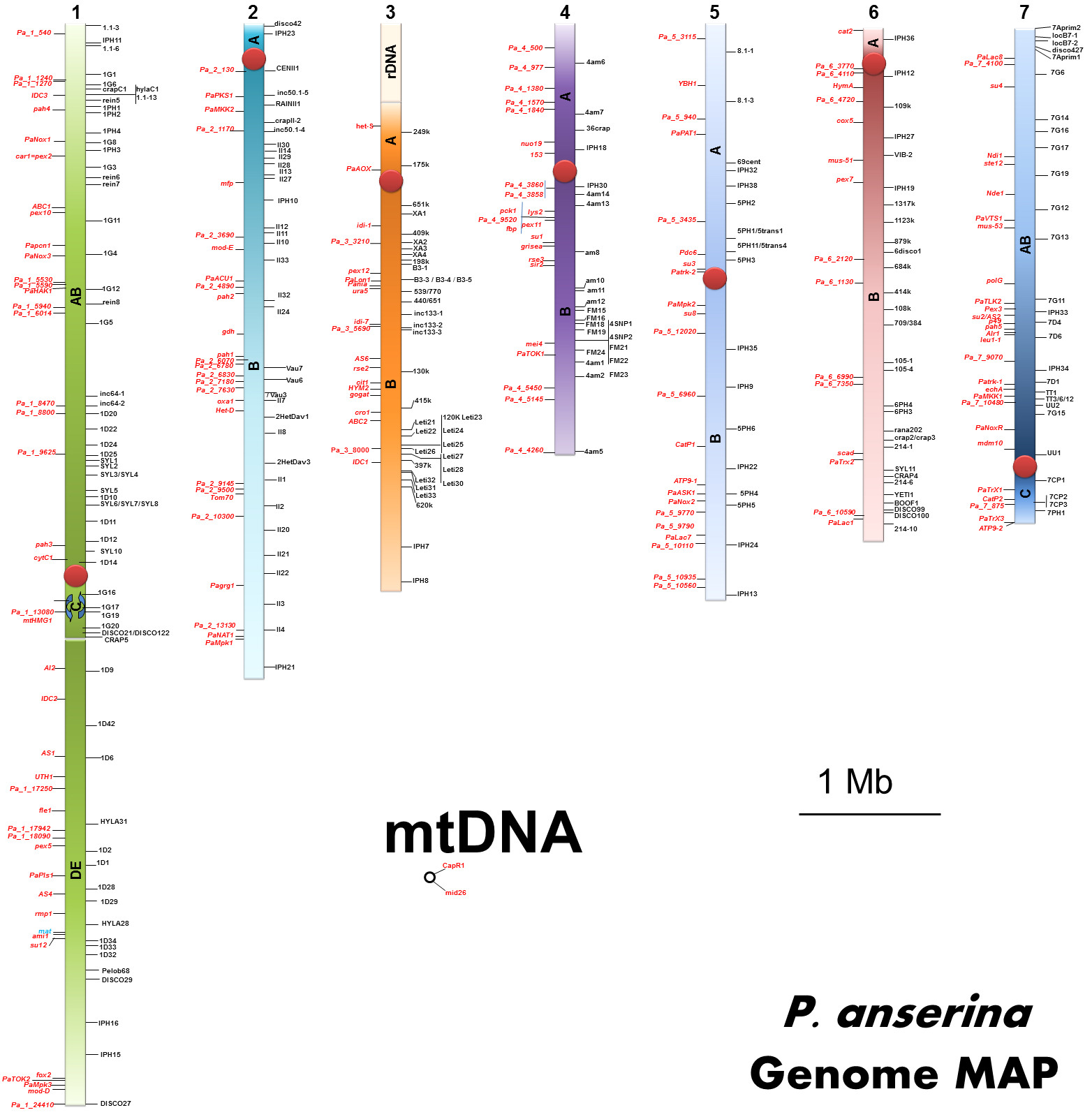 |
A finishing of the genome of strain S mat+ is now underway. Génoscope has produced a new assembly with a 20X coverage of 454 sequences. The successive Releases presented on this site stem from a combination of the old and new assemblies as well as manual targeted gap filling by PCR.