Podospora anserina Genome Project
- How can I cultivate Podospora anserina in the lab?
- Strains preservation
- Genetical analysis
- Transformation
- DNA, RNA and proteins extractions
How can I cultivate Podospora anserina in the lab?
In nature, Podospora anserina grows on herbivores dung. In the lab, the fungus is cultivated on well-defined synthetic media allowing for the wild-type strain to grow in optimal conditions.
Several synthetic media are routinely used. Each one has its own special usage:
M2 Medium is the most commonly used medium. Crosses, Stocks and most phenotypic tests are made on it. It permits the mycelium to grow at about 7mm per day. In stationary phase (photo here), the mycelium 1) develops aerial hyphae, 2) presents a green pigmentation, and 3) accomplishes sexual reproduction in a week (4 days between fertilisation and ejection of the ascospores out of the perithecium). The longevity experiments made on Podospora anserina strains are performed with glass tubes, called race tubes, instead of Petri plates. The medium in race tubes contains a double amount of agar |
M2 medium Composition: KH2PO4 0,25 g/l K2HPO4 0,3 g/l MgSO4 0,25 g/l (if MgSO4 7H2O) Urea 0,5 g/l Thiamine 0,05 mg/l Biotine 0,05 µg/l Citric Acid 5 mg/l ZnSO4 5 mg/l CuSO4 0,25 mg/l MnSO4 50 µg/l Boric Acid 50 µg/l Natrium Molybdate 50 µg/l Alun de Fer 1 mg/l Dextrine 5,5 g/l dissolved in warm water Agar 10 g/l Adjust to pH7 with a KH2PO4 solution. Autoclave for 20 minutes at 120°C |
MR Medium is a semi-defined medium that is mainly used to recover the mycelium. To this end, explants are inoculated on a MR dish recovered with a sterile cellophane sheet. In these conditions, the mycelium regenerates and grows on the cellophane layer without invading the medium. The mycelium is then easily recovered by simply scraping the cellophane sheet. On this medium, the hyphae develop no or few aerial mycelium (photo here). |
MR Medium composition: Corn flour 25 g/l Corn cream 25 g/l Dissolve with H2O Mix and incubate overnight at 58°C Filter the mix twice, then adjust with H2O again Agar 12 g/l Autoclave for 20 minutes at 120°C |
RG Medium is an hyperosmotic medium mainly used for protoplasts regeneration (after mutagenesis or transformation). |
RG medium Composition: Same as M2 + 205 g/l of saccharose Autoclave for 20 minutes at 120°C |
G medium is mainly used for ascospore germination. It allows the germination of nearly 99% of wild-type ascospores in. On this medium, the strains lack pigment (photo here). |
G medium Composition: Same as MR + 6 g/l Ammonium Acetate Autoclave for 20 minutes at 120°C |
Sorbose Medium is only used to obtain male gametes (microconidia) in high amount. To this end, the sorbose medium is poured in tilted assay tubes. Numerous explants are then innoculated in these tubes. After growth, one must add 2 ml of sterile water, vortex and filter the suspension with sintered glass filter n°4. |
Sorbose medium Composition: Yeast Extract 2 g/l Sorbose 2 g/l Glucose 1 g/l Agar 12 g/l Autoclave for 20 minutes at 120°C |
Agar covers are used to recover ascospores that are ejected outside the perithecia. Ascospores on these covers can resist for a few weeks before dying. |
Agar covers are made by pouring a thin layer of melted Agar in a Petri plate at the concentration of 80 g/l supplemented with NaCl at 20 g/l (autoclave for 20 min at 120°C and pour very hot). |
Supplements : In these media, it is possible to add various metabolites or antibiotics to select special strains or to allow auxotrophic strain to grow. |
Uridine 100µg/ml Leucine 50µg/ml Lysine 100µg/ml Tryptophane 30µg/ml Methionine 30 µg/ml Antibiotics Hygromycin B 75µg/ml Paromomycin 500µg/ml Phleomycine 10µg/ml |
Strains preservation
There is not problem for long term conservation of P. anserina strains. Several strategies are available.
1- at 4°C
Put several explants on a M2 plate. Incubate until the plate is covered with mycelium. Conserve the stock at 4°C with a parafilm wrap. These stocks can be conserved for at least 2 years.
2- at 80°C
Put several explants on a M2 plate After 2-3 days of growth, take numerous large explants at the growing edge. Store them in a tube containing liquid RG (i.e. RG without Agar). Place directly the tubes at -80°C. These tubes can be frozen/defrosted numerous times without notable effect on explants regeneration. The strains can be conserved in this way for several years (in the lab, some strains are frozen for more than 8 years and the explants still regenerate without any problem).
Note that some mutant strains may not be stocked as well.
Genetical analysis
This page deals only with advance genetics. If you are not familiar with genetics, consult a textbook !
1- Mutagenesis
UV mutagenesis at 254 nm is the most convenient method. Doses for wild type range from 100 J/m2 to 300 J/m2, usual dose being 200 J/m2. It corresponds to 1% survival when measured with protoplasts regenerative potential. Screens obviously depend upon the desired mutants. However, if no mutants are obtained with a direct screen there is still the possibility to do mutagenesis on protoplasts. To do this, one needs to do protoplasts as indicated below, then spread them on RG medium at the concentration of 104 per plate. Irradiate with UV at 200J/m2. Incubate in the dark at 27°C. Regenerants can be picked up after 48h under the binocular to test them individually. At least 5000 of them should be scrutinised before giving up !
2- Dominance and recessivity
Podospora anserina has a strictly haplobiontic life cycle and vegetative diploids are difficult to select for. So, apart from the mutations that act between fertilisation and meiosis, tests must be performed on heterokaryons. To obtain heterokaryons, there are several simple possibilities : - you can mix explants from both mycelia by simply grinding them in an eppendorf tube with a yellow tip and then inoculate the mixing on a new plate. - or you can obtain directly after sexual reproduction the heterocaryotic strain as indicated here.
Note that these tests in heterocaryons can yield some interpretation problems since very often mutant nuclei (sometime wild-type ones) can be easily lost. To circumvent this there are simple genetics tricks:
- one can use strains with complementary mating type, allowing to check (through the presence of perithecia) if both nuclear types are present.
- one can use the 193 mutation, that affect ascospore and mycelium colour (wild type is green, mutant is white). This can be used to test strains that do, not differentiate perithecia. Heterocaryotic strain with the mutant and 193 will yield green perithecia only if the mutant is recessive, perithecia should be absent or pink if the mutation is dominant.
- It is possible to associate your mutation with the leu1-1 (or lys2-1) mutations and to make a balanced heterocaryon with lys2-1 (or leu1-1). Growth on minimal medium will ensure that both types of nuclei are present.
Note that a problem frequently encountered in heterocaryon construction is the "vegetative incompatibility". Indeed, when anastomoses occur with culture of the same strain, heterocaryon will readily be formed but when anastomoses occur between different wild-type strains, frequently hyphae will fuse but die in a violent reaction called incompatibility reaction. In summary, it is advised to do heterokaryon with strains having the same genetical background.
3- Complementation
Complementation tests are carried out with heterokaryons as described in the above section.
4- Segregation
Modalities of segregation in Podospora anserina depends on its life cycle, which is described here.
Note that for a better understanding of the process, it can be summarised as follows in a cross a- x a+:
| first meiotic division without a crossing-over between a and the centromere | 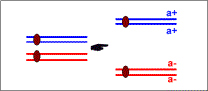 | first meiotic division with a crossing-over between a and the centromere |  |
| second meiotic division |  | second meiotic division | 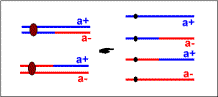 |
| postmeiotic mitosis in which spindles are orthogonal to the meiotic spindles |  | postmeiotic mitosis in which spindles are orthogonal to the meiotic spindles | 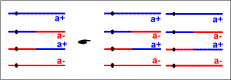 |
| delimitation of the 4 ascospores around two non-brother nuclei resulting from postmeiotic mitosis | 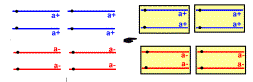 | delimitation of the 4 ascospores around two non-brother nuclei resulting from postmeiotic mitosis | 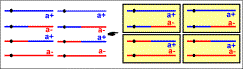 |
Prominent features of Podospora anserina ascus analysis are :
1°) Ascospores are primitively dikaryotic. However, before ascospore germination nuclei multiply and a ripe spore contains around 30 nuclei. This allows for the recovery of heterocaryotic strains for dominance/recessivity and complementation tests.
2°) Postmeiotic mitosis spindles orientation entails that (1) if no crossovers occurred between the gene and its centromere, the ascus contains two ascospores with two a- nuclei and two with a+ nuclei; (2) if a crossover has occurred between the gene and its centromere, the ascus contains four ascospores with one a- nucleus and one a+ nucleus. First (FDS) and second (SDS) segregation frequencies can be easily obtained. Note that this allows to do ordered tetrad analysis with disordered tetrads !
3°) In about 1% of the ascus, ascospore delimitation is not usual and two "small spores" are created instead of a "big one". This permits the easy recovery of homocaryotic strains.
4°) Crossing-over are not randomly scattered and there is a strong interference phenomenon. Especially, in 98% of the meiosis, a single crossing-over occurs between the mating type and its centromere, allowing for the recovery of self-fertile mat+/mat- strains. To see the genetical map of Podospora anserina, click here.
5°) It is possible to do male or female oriented crosses to check for cytoplasmic segregation.
To proceed, one must set up the desired cross on a Petri plate that contains M2 medium by inoculating 4 explants of each strain 1 cm apart. If a directed cross is not required, wait. If a directed cross is required, after 3 days pour 1.5 ml of water and spread the conidia (those will disperse because they are not tightly attached to the mycelium whereas ascogonium are not so easily removed); then remove the male thallus with a scalpel so that perithecia develop only on the female side. Perithecia will produce ascospore about 4 days after fertilization and do so for 3- 6 days. Ascospores are recovered on Agar plates put upside down on the cross plate. Ascospore can be recovered with a needle glued on a stick under a binocular (try again, at first it may be difficult !) and inoculated on a G medium Petri plate. Percentage of germination is nearly 100%. After two days the thalli have a diameter of 1 cm and can be sliced to test them.
Transformation
Protoplasts are prepared for transformation according to the following procedure. All steps are performed in sterile conditions.
- Roux flasks are inoculated with mycelia fragmented in a Waring blender for 15 sec.
- Cultures are grown for 30 h at 27° C in the dark in liquid minimal medium supplemented with 2.5 mg/ml yeast extract.
- Mycelium is harvested on cheesecloth and washed with TPS1 buffer (0.6 M saccharose, 5 mM Na2HPO4, 45 mM KH2PO4).
- Weight the mycelium. X g of wet mycelium is put in X ml of TPS1 containing 40 mg/ml of Glucanex (Novo Nordisk Ferment AG) and digested for 2 to 3 h at 37° C.
- The protoplasts are separated from mycelial debris by filtration through cheesecloth.
- They are concentrated by 10 min centrifugation at 3200 rpm, washed twice with TPS1 and once with TPC buffer (0.6M saccharose, 10 mM CaCl2, 10 mM Tris, pH 7.5).
- The final pellet is resuspended in TPC buffer and protoplasts concentration is determined by counting under microscope with a hemacytometer. Protoplasts can be transformed immediately or stored at - 70° C.
- Before transformation, the protoplasts are subjected to a 5 min heat shock at 48° C and transferred on ice.
- The DNA is added (5 microg of DNA for 0.2 ml of protoplasts at a 108/ml concentration) and protoplasts are incubated for 10 min at room temperature.
- Two ml of a PEG solution (60% polyethylene glycol 4000, 10 mM CaCl2, 10 mM Tris pH 7.5) are added and carefully mixed.
- After a 15 min incubation, the protoplasts are distributed in tubes containing top agar placed in a 42°C water bath and plated on minimal selective medium containing 0.8 M saccharose as osmotic stabilizer (top agar contains 0.2M sorbitol in addition to saccharose).
Routinely 10 to 50 transformants are obtained per microg of plasmid DNA. Plasmids carrying different selective markers are available for transforming Podospora anserina protoplasts. The leu1 or ura5 wild-type genes restore growth on minimal medium when introduced into the mutant recipient auxotrophic for leucine or uridine, respectively. The hph gene or the ble gene allow selection of transformants resistant to hygromycin (100 microg/ml) or to phleomycin (5 microg/ml), respectively.
Plasmids sequences are available, here : pBhyg, pBC-hygro, and pBC-phleo.
DNA, RNA and proteins extractions
Several methods are available to extract macromolecules from Podospora anserina mycelium. Here are a few that should work.
1- DNA Extraction. Large scale prep.
- cultivate the mycelium in Roux flasks (M2 supplemented with 10g/l of difco yeast extract). Filter to recover the mycelium
- freeze in liquid nitrogen
- grind it with a mortar
For 2 g of mycelium
add 10 ml of ice cold TES/Sarkosyl (Sarkosyl 2,5%, tris 12,5 mM, EDTA, 12,5 mM, NaCl, 25 mM pH=8) made at the last minute from stocks of Sarkosyl 5% and TES 4x
- incubate for 1h30 at 4°C mix frequently
- centrifuge for 10 min at 6000 rpm, 4°C
- dialyse supernatant against TES 1x at 4°C overnight
- do alpha-amylase treatment for 2 hours at 20°C (500 ml added from a solution at 2,5 mg/ml in NaCl 3,5M kept at -20°C)
- do proteinase K treatment for 2 hours at 20°C (50 ml added from a solution at 20 mg/ml kept at -20°C)
- add 2 vol ethanol 100° and 1/10 vol sodium acetate 3M pH=6
- centrifuge for 5 min at 8000 rpm
- recover in water to make a cesium centrifugation:
for a 65VTI rotor, add 4ml sterile water, 4,6 g of cesium chloride and 20 ml de DAPI 1mg/ml, centrifuge for 12h at 45000 rpm
This method allows the separation of nuclear (lower band) and mitochondrial (upper band) DNA.
yield is around 1 mg/bottle
2- DNA extraction Small scale prep.
- inoculate a MR Petri plate containing a sheet of sterile cellophane. let grow for 2-3 days
- recover the mycelium with a spatula and put it into a 2 ml eppendorf that contains 600 ml of TNE/SDS (Tris 10 mM, EDTA 1 mM, NaCl 100 mM and SDS 2%) to be made from stocks of SDS 10% and TNE 2X
- do 3 cycles of freeze/thaw in liquide nitrogen /water bath at 70°C; vortex between each cycle
- add one volume of phenol pH=8
- centrifuge for 5 min at 4°C 13000 rpm and retrieve the supernatant
- repeat with phenol + chloroform and with only chloroform
- precipitate the supernatant by adding 2 vol ethanol 100° and 1/10 vol sodium acetate 3M pH=6
- rinse with ethanol 70°, dry and suspend in 40 ml of sterile water
yield : a few mg/plate
3- RNA Extraction
- Recover the mycelium (from Roux flasks for large scale preps or MR plates + cellophane for small scale preps)
- freeze in liquid nitrogen
- suspend in 1 or 2 vol of hot phenol made by incubating at 70°C a mix of 1 vol phenol pH=7,5 and 1 vol of tpRNA (NaCl 100 mM, Tris 10 mM pH=7,5, EDTA 1 mM, SDS 0,1%, Sarkosyl 2%), the mix must be homogenous
- incubate at 70°C for 5 min
- centrifuge for 5 min at 14000 rpm for small scale preps and 8000 rpm for large scale preps
- recover the supernatant in a new tube
- repeat by adding phenol + chloroform and then only chloroform
- add 2 vol. of LiCl and let precipitate overnight at -20°C
- centrifuge for 5 min at 14000 rpm for small scale preps and 8000 rpm for large scale preps
The RNA is very clean and can be used for all usage (northern, cDNA, RT-PCR ...)
4- Protein Extraction and dosages
- Do 36 hours cultures and recover the mycelium
- grind it in a mortar and in liquid nitrogen
- the powder is suspended in the following buffer (10mM Tris pH7.5, 1mM EDTA, 76mM glycine)
- centrifuge for 10 min at 11000 rpm at 4°C, the supernatant is recovered and kept on ice
- dosages are made directly on the supernatant, treated or untreated with triton 2%